Lab 10: SDMs: Modeling Methods
This lab is a continuation of species distribution models (SDMs) using the R package, dismo: https://cran.r-project.org/web/packages/dismo/index.html. The package contains a great vignette and you worked through the first part of it in Lab 8, and the second part in Lab 9. You should complete Lab 8 & 9’s portion of the vignette with your species of choice before starting on Lab 10.
For this lab, you will work through Part 3 (Chapters 8-13) of the dismo vignette. You can find the PDF of the vignette here: sdm.pdf, and as HTML here: https://rspatial.org/raster/sdm/index.html.
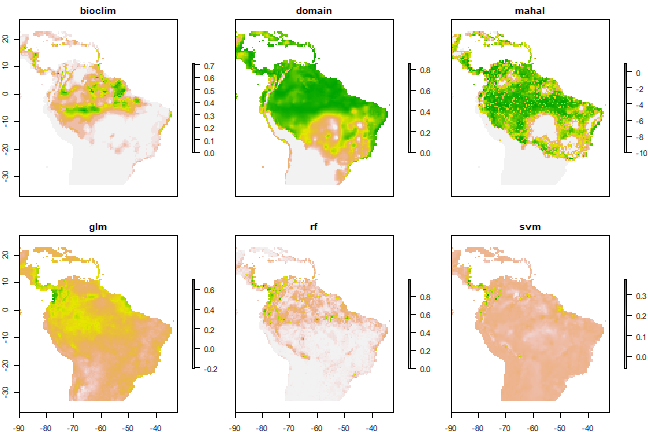
Comparison of outputs for various SDM algorithms from the dismo vignette.
Types of SDM Models
In Lab 8, you filled in a template describing a particular type of SDM of your choice. See D2L for those summaries as a reference.
For this Lab, work through Ch. 8-13 (from Modeling Methods through Geographic Modles) with your species’ data from Lab 9. Get as far as you can by building multiple models with different algorithms, and comparing them. Working on the Geographic Models (Ch. 13) is optional.
After completing the vignette with your species of choice, answer the following QUESTIONS:
QUESTION 1:
Describe the differences you observe in the mapped prediction between the different modeling algorithms. Which mapped prediction is closer to the known distribution of the species? Provide a source and image (e.g., screenshot) of an independent source showing the range map of the species.
QUESTION 2:
Of all the models you tried, which model performs best for your species’ data? Why? Include plots and model statistics to help with your explanation. Do you agree that model averaging (i.e., ensemble modeling) is superior to selecting one top model? Why or why not?

This work is licensed under a Licensed under CC-BY 4.0 2020 by Phoebe Zarnetske.